How to Upload Data
Synapse.org is the data management software for all data hosted and shared on the NF Data Portal. This means that you will upload data to be stored in Synapse, and it will be ultimately be shared on the NF Data Portal. Think of Synapse as the back-end and the NF Data Portal as the front-end for your data.
You will be uploading data into Synapse using a variety of tools. This article will give you a brief guide to ensure you upload data to the correct place, using the appropriate methods.
At the end of this page, you’ll find a video tutorial that covers the steps involved in uploading data (steps 3-4 below). This is the second in a series of three videos on sharing data. Find the first video, Introduction to Sharing Data (steps 1-2), here.
Uploading data involves the following actions, to be completed in order:
1. Ensure your account is certified
If your account is certified, there will be an icon indicating so on your profile page. If it is not yet certified, see Step 1 in How to Share Data (an Overview) for instructions.
2. Locate your Synapse project
In order to upload data, you need to have a designated Synapse project, which will be provided to you once you register your project with the NF-OSI coordinators. If you haven’t registered your project yet, see How to Share Data (an Overview) for more information and instructions on how to do this.
If you have successfully registered your project, you should have been provided a Synapse project space to house your data. Here’s how to find this:
Log into Synapse.org
Click on your profile picture in the upper right hand corner and click Projects
Click on your project—the project title should match the information you provided the NF-OSI coordinators when you registered your project
3. Create a folder for your data (if not already set up)
In most cases, your dataset-specific folders should already be set up based on the datasets listed in your data sharing plan. However, here are the general instructions for your creating data folders:
Once you’re in your project, click on Files, and then click the Data or Raw Data folder.
Navigate to, or create a folder (Folder Tools > Create Folder) for the experiment or data type you plan to upload (e.g. “RNA Seq Experiment 1”).
You can find more information on Synapse projects, files, and folders on the Synapse docs site.
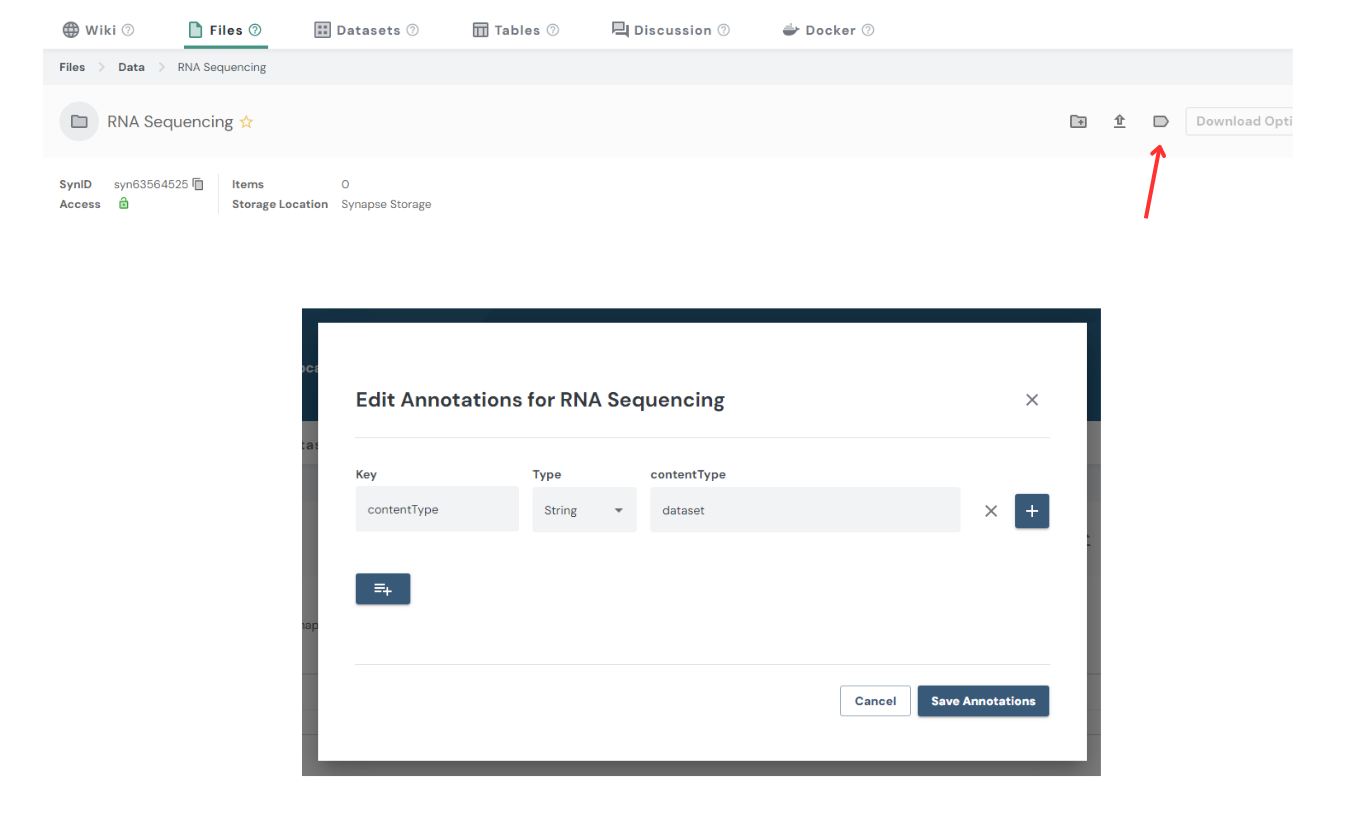
Important: If you manually create your folder, the folder will not show up in the Data Curator App unless there it has the annotation contentType = dataset. You can manually add this annotation by clicking the annotation folder icon (shown below by the red arrow) and adding the Key contentType to equal dataset. Folders created by Sage will have this annotation automatically.
4. Upload your data
Now that you have an appropriate folder to house your data, you’re ready to upload. You can do this using the Synapse.org UI or by using one of our programmatic clients—Python, R, or command-line. For small folders/files, the website upload is generally easiest. For larger uploads (large files or many files), we recommend using the programmatic clients.
To upload using the web UI:
navigate to the folder you want to upload to and click Folder Tools → Upload or Link to a File.
To upload using the Python client:
To upload using the command-line client (requires the Python client):
To upload using the R client:
follow the instructions on our R docs site, or
follow the bulk-upload instructions to upload using a manifest on our Synapse docs site
To upload files that already exist in an S3 bucket or Google Cloud Storage Bucket:
As an alternative option, if your files are already on GEO, SRA, or another platform that doesn’t require a login, you can link directly to the download HTTPS or FTP URL (for example, the links under the Data Access tab on this SRA page). Here’s how:
in your Synapse account, under Folder Tools, click Upload or Link to a File
simply add in the link
Using this method, users will be able to download the file as if it lived on the NF Data Portal/Synapse, but you only need to upload it once! Please note that annotation on Synapse is still required with this method.
If you have any questions about whether your repository of choice is compatible with this, please reach out to us (nf-osi@sagebionetworks.org).
6. Annotate your data
Once your dataset is uploaded, you must annotate it in order to make it findable and understandable. You can find those instructions at How to Annotate Data.
Video Tutorial: Uploading Data via the Synapse UI
Links mentioned throughout the video:
